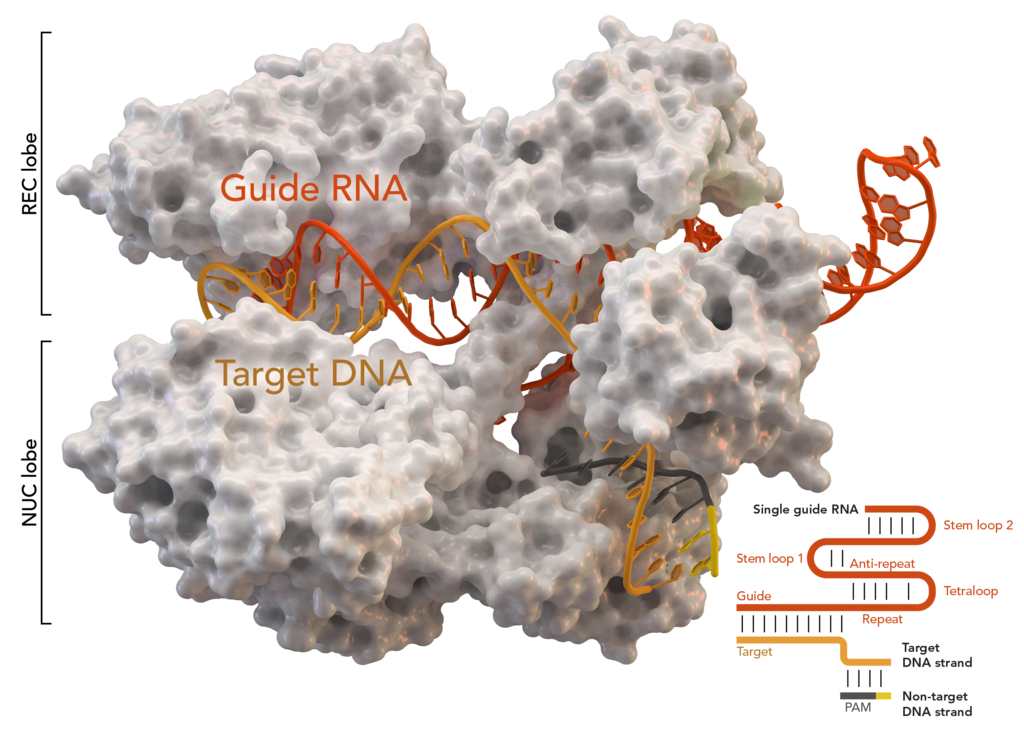
Crystal structure of S. aureus Cas9 in a complex with crRNA and its target DNA
In previous posts you asked to tell in more detail about CRISPR / Cas9 and approaches to genome editing. I just did not include such a voluminous material in the last post. And in this I will not include, for example, the TALEN method, which can potentially be more accurate and has its advantages. But darling, yes.
Let's try to walk through the already well-described method of genetic modification using CRISPR-Cas9 and look a little more broadly at the prospects that it opens up for us. I was wildly interested in the direction of xenogeneic transplantation from pigs to humans. The first, "pre-correction" methods showed that such a pig transplant is killed by the body within a few minutes . But the method was not discarded at all. Still, this is a promising way not to wait until another potential organ donor dies, but to grow them in advance. Then it turned out that pigs also carry a pack of specific retroviruses that are built into their genome and can cause an epidemic of a new xenozoonosis. And viruses, too, must somehow be picked out before transplantation. And somewhere here people in white coats appear on the stage, doing something incomprehensible in their laboratories ...
The bacterial antivirus that turned everything upside down
In 1987, CRISPR was discovered by Japanese scientists led by Yoshizumi Ishino. At that time, they drew attention to the unusual repeats in the E. coli genome, but did not attach particular importance to this. And only when similar repeats were found in archaea, which are genetically very far from E. coli, did they begin to search for similar structures in other prokaryotes. These snippets are called CRISPR - clustered regularly interspaced short palindromic repeats. A little later, special CRISPR-associated proteins, Cas ( CRISPR as sociated protein), were also discovered. Well discovered and discovered, it would seem. But they were the beginning of a new biotechnological revolution.
Now I will try to tell you how it works and why bacteria need it. The bacteria would love to survive. But they like to parasitize viruses. When someone eats you - it's unpleasant and you have to do something. As a result, the answer was a complex but extremely interesting mechanism. The bacterium tremulously stores in a piece of its genome the signatures of all the villains who are trying to infect it. These are the same repeating palindromic regions - CRISPR. They work together with proteins from the Cas group. We are interested in the key Cas9. How does it look live, if simplified a little?
If this system does not work, then the bacterial cell is unable to distinguish the viral genome, carefully introduced by the bacteriophage, from its own. And the not very clever system of protein synthesis is immediately reoriented to the release of new generations of phages. The cell dies.
If CRISPR / Cas9 is triggered, then the process is different. The bacterium uses the data recorded in CRISPR to create control RNA. The protein complex begins unwinding the DNA for testing before "launch". In case the sequence matches the viral signatures recorded in CRISPR, Cas9 raises the alarm and immediately cuts off the recognized enemy fragment. That is, even if a virus is embedded in the bacterial genome at the “firmware” level, it will be cut out from there as soon as the built-in antivirus recognizes it.
Nobel Prize
Emmanuelle Charpentier and Jennifer Doudna received the 2020 Nobel Prize in Chemistry for the creation of new technologies that allow genome editing using CRISPR-Cas9. This technology made it possible to replace the more accurate at that time, but more complex and expensive methods of zinc fingers and TALEN nucleases. Past methods required the development, expression and validation of a completely new pair of polypeptides for each new target locus. And CRISPR-Cas9 gave a standard tool, when used in the minimal version, it is enough to obtain the necessary control sequence, according to which Cas9 will find the area necessary for dissection.
Okay, we've cut DNA from a human, yeast, or a test rat. This is cool, but now we have two broken pieces and a destroyed chain. There are several options for stitching back.

You can try to match the cut pieces using the non-homologous end-joining method. He's on the right. To put it simply, in this version we are simply trying to dock the double break directly and weld it into a single whole. This mechanism is rather ineffective, in the process of "fitting" individual end portions may fall out. As a result, small fragments are often lost in the break zone or, on the contrary, short inserts appear. This approach usually turns off the gene irreversibly, rendering it defective.
The second option is more interesting. He is in the illustration on the right. Repair by homologous recombination implies the replacement of a deleted sequence with a new sequence complementary to a repair template created by the researcher himself. As a result, it is possible not only to turn off the gene somehow, but to replace the mutant sequence with the normal one.
The main problem with the method is that it is probabilistic. Yes, in a large percentage of cases, it will work exactly as it should. But in many cells, it will either not give the desired effect, or break something to hell. And it's good if just single cells die from this, and do not become, for example, tumor cells. Therefore, all such changes must be thoroughly tested. Fortunately, relatively fresh approaches to improve the specificity by creating a custom Cas9 can reduce the number of erroneous cuts to almost zero.
Xenografts
As I said before, the massive use of these techniques stops specificity. If our wonderful molecule just misses 20% of the cells, it's not a big deal. This will mean that 80% of human cells with a congenital mutation are fixed and will already begin to produce the correct enzyme, divide adequately, or do something else correctly. As a rule, this is more than enough for a person to become clinically healthy.
In this section, I would like to talk a little about how genome editing can potentially solve the problem of organ donation. I must say that the lack of organs for transplantation looks very sad. Huge waiting times and a lot of ethical issues.
The number of organ transplants performed in Russia is hundreds of times lowerneeds. If the same kidney can be transplanted from a suitable relative, then a heart transplant is already guaranteed to mean the death of the donor. One of the key areas in the search for an unlimited source of organs is xenotransplantation. This is an option for transplanting tissues and organs between different species. One of the most suitable donors in structure and size for humans is a pig , oddly enough. Primates, although genetically closer, are generally much smaller in size and very expensive to breed. Unfortunately, early experiments with pig organ transplants showed that they begin to reject with a hyperacute response within a few minutes after being connected to the bloodstream.
How to modify a pig

In order for the transplant not to be rejected, it is necessary at least to knock out the target genes responsible for the synthesis of the most foreign proteins to us. The first such protein is the enzyme alpha-1,3-galactose, which in all primates is irreversibly broken in the course of evolution. It is he who causes the onset of rejection within a few minutes . Gene correction allowed to create a breed of pigswith a switched off gene responsible for the synthesis of the enzyme - pig GTKO. Although this significantly slowed down the rejection process, it did not completely stop it. It turned out that N-glycolylneuraminic acid and β1,4 N-acetyl galactosaminyltransferase, which are absent in primates, are also problematic. These genes were also knocked out and received the pigs GGTA1 / CMAH / β4GALNT2 KO with all three genes turned off at once. Presumably, this can practically neutralize the rejection reaction . In theory, if it is possible to also force pig cells to synthesize the surface glycoprotein CD47 of a person, then the compatibility will be quite excellent .
There was a very promising publication in 2018 on heart transplantation from GMO pigs to baboons. Of the 5 baboons, only one had problems, pulmonary edema began for surgical reasons and had to be withdrawn from the experiment ahead of time. The rest survived until the end of their experiment period with good health. Two to three months and two to six months. There were some problems due to the growth of the organ, since a pig's heart is larger than a baboon's, but for a person this will not be a problem.
Is there a new HIV ahead of us?

Phylogenetic tree of HIV and related viruses in chimpanzees
It would seem that a bright future awaits us, where a separate herd of special laboratory pigs will graze for the production of donor organs. But here, too, a lot of ethical problems emerge. I suspect that such a method will be unacceptable for representatives of several religions, which automatically makes it inaccessible to a large proportion of the world's population.
Pigs also have retroviruses. These include, for example, such a wonderful lentivirus as the human immunodeficiency virus and related primate viruses. It is believed that it was the primate virus that gave rise to the massive HIV epidemic in the 70s of the last century. And now a person does not know what to do with him. Pigs are carriers of PERVs - porcine endogenous retroviruses. And you cannot get rid of these viruses by careful breeding and anti-epidemic measures, since the viruses are already firmly embedded in the genome of their cells. Research was conducted which should give a possible solution. To begin with, they took pig cells and grew them in the same culture as embryonic cells from a human kidney. The most annoying thing is that they were able to identify human cells infected with porcine retroviruses, which potentially indicates such a possibility for transplantation. As a solution, they proposed pre-processing of cells with CRISPR-Cas9 and complete excision of all viral fragments from the culture of porcine renal epithelium cells.
Biotech and risks
Where all this will lead is difficult to predict. Unfortunately, the risks of getting an exotic viral mutant from such a transplant are far from zero. And there is a chance that such an infection can also spread quietly and imperceptibly, before it becomes obvious, as happened in its time with HIV. And yet I am rather optimistic. We will inevitably step on a lot of rake in the process of research, but now we have the tools to treat previously fatal diseases and prolong human life.
