Training relevance
An important stage in the development of the classification of the order of fleas is associated with the development of genomolecular research. At present, thanks to these methods, the kinship of the orders of fleas and mecopter is confirmed. The genes of ribosomal DNA, the elongation factor of protein synthesis and the mitochondrial gene cytochrome oxidase II (COII), which have been studied in representatives of most families of the mecopter order, as well as 128 species of 83 genera of fleas from 16 families, are being actively studied (S.G. Medvedev. 2009; Whiting 2002; Whiting et al. 2008). Flea phylogeny is reconstructed on the basis of data on these genes by various methods of constructing phylogenetic trees and models for calculating pairwise distances, which is often presented in the form of a consensus tree (strick consensus) (Whiting et al. 2008). However, in some cases, the results of genomolecular studies contradict the scheme of family ties,obtained on the basis of phylogenetic analysis. In addition, in accordance with different interpretations of molecular genetic data, the origin of the key infraorders of fleas can be interpreted in different ways. So, from the point of view of different authors, the same clades of fleas can be both polyphyletic and monophyletic groups (Whiting et al. 2008, S.G. Medvedev. 2019). This may be due to both the peculiarities of the ecology of the modern flea fauna and various methodological problems of working with molecular genetic data.and monophyletic groups (Whiting et al. 2008, S.G. Medvedev. 2019). This may be due to both the peculiarities of the ecology of the modern flea fauna and various methodological problems of working with molecular genetic data.and monophyletic groups (Whiting et al. 2008, S.G. Medvedev. 2019). This may be due to both the peculiarities of the ecology of the modern flea fauna and various methodological problems of working with molecular genetic data.
Regarding the first point, it should be emphasized that the isolation of individual phyletic lines of fleas, which have an equal status in taxonomy, occurred at different historical intervals. This, in turn, means that the flea fauna is scattered fragments of an extensive paleofauna, which is largely extinct by now (S.G. Medvedev. 2009, S.G. Medvedev. 2019). It follows from this that with different methodological approaches to genetic molecular studies, we can lose sight of the convergent evolution of nucleotide sequences (homoplasia), which can lead to errors in determining the topology of the tree and to low statistical support of branches. So, for example, using data that have different relationships for different taxa and, accordingly, different relationships to different regions of genes,we will find gaps in the sequences, and the absence of data for different loci for different samples (S.G. Medvedev. 2019, Lukashov V.V. 2006). Therefore, when choosing a research method, all these problems should be taken into account and samples of the most studied and common flea taxa should be selected in order to then compare them with the most studied mecopter samples. This way we can build a more reliable tree with high support.
, Siphonaptera Mecoptera -II (COII).
:
Siphonaptera NCBI [5], .
(COX 1) Siphonaptera .
-II (COII) Mecoptera Siphonaptera NCBI [1].
MEGA X.
.
NCBI [5] : Chaetopsylla globiceps (Taschenberg, 1880), Chaetopsylla trichosa (Kohaut, 1903), Ctenocephalides felis (Bouché, 1835), Ctenocephalides canis (Curtis, 1826), Ceratophyllus sciurorum (Schrank, 1803), Leptopsylla segnis (Schönherr, 1811). , NCBI[1], Boreus: Boreus westwoodi (Hagen, 1866), Boreus hymalis (Linnaeus, 1767), Boreus coloradensis (Byers, 1955), Boreus nivoriundus (Fitch, 1847), Boreus brumalis (Fitch, 1847), Boreus californicus (Packard, 1870), (.1.).
UPGMA [2] , - 500 [3]. Muscle (. 2.) MEGA X [4]. Muscle . , . translate DNA to protein and vice versa (.3), . (. 3.) , - , . (.4). MEGA . Modeltest 3.06 PPC (Posada, D., Crandall, K.C., 1999) (AIC), . AIC , , (Posada, D., Buckley, T.R., 2004.). MEGA X [4] 64- Windows 10 Home 4- INTEL 1,6 1 .
- [6] (.5.). 12 . 1-+2-+3-+. ( ). 742 .
( 1.)

(.2.)
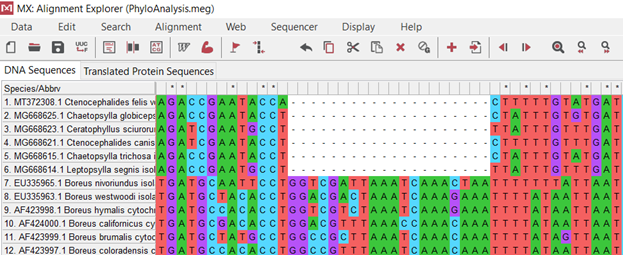
(.3.)
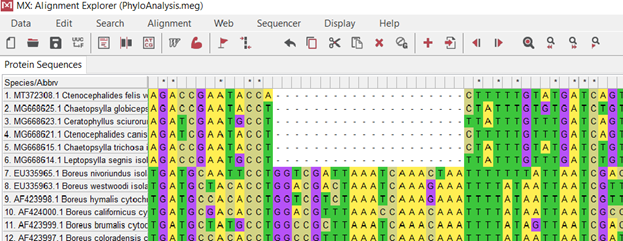
(.4.)
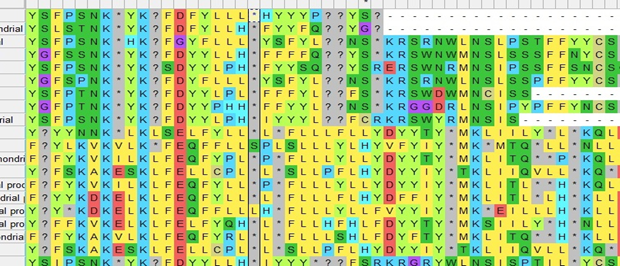
(.5)
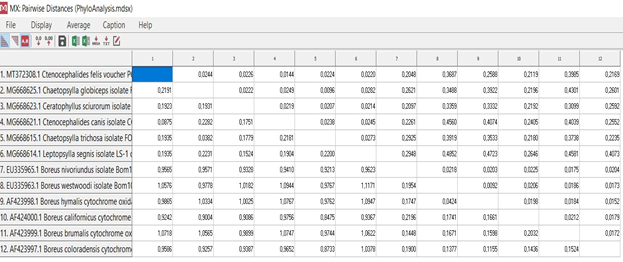
. Mecoptera Siphonaptera. 60% , Mecoptera Siphonaptera. , , , .
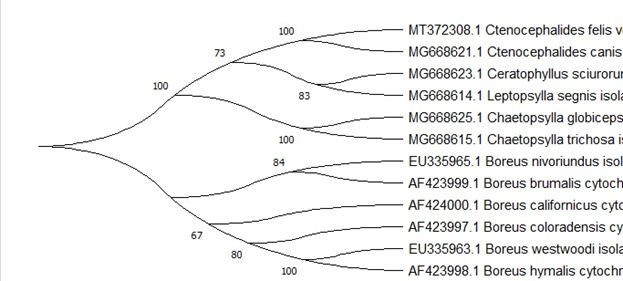
, MEGA X [4] (80%-100%) , . Ceratophyllus sciurorum Leptopsylla segnis (73%), . , (.. .2009; Whiting 2002; Whiting et al. 2008).
Mecoptera Siphonaptera . . Boreus nivoriundus Boreus brumalis (80%) «» Chaetopsylla globiceps Chaetopsylla trichosa (100%). , Mecoptera Siphonaptera. Boreus californicus (67%) . , . (.6.)
(.6.)
. . , , , . . . . [https://vk.com/phanerozoi] . , .
.
, , .
, .
1. Whiting M.F. 2002. Mecoptera is paraphyletic: multiple genes and phylogeny of Mecoptera and Siphonaptera. Zoologica Scripta, 31: 93–104
2. Whiting M.F., Whiting A.S., Hastriter M.W. and Dittmar K. 2008. A molecular phylogeny of fleas (Insecta: Siphonaptera) and host associations. Cladistics, 24: 1–31.
3. , .. , 313, № 3, 2009, c. 273–282
4. (SIPHONAPTERA) .., .. III , 2009, . 185-190
5. / .. —.. , 2009. — .256. .29-31.
6. Posada, D., Buckley, T.R., 2004. Model selection and model averaging in phylogenetics: advantages of the AIC and Bayesian approaches over likelihood ratio tests. Syst. Biol. 53, 793–808.
7. Posada, D., Crandall, K.C., 1999. Modeltest. Bioinformatics 14, 817–818.
1. http://lifemap.univ-lyon1.fr
2. Sneath P.H.A. and Sokal R.R. (1973). Numerical Taxonomy. Freeman, San Francisco.
3. Felsenstein J. (1985). Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39:783-791
4. Kumar S., Stecher G., Li M., Knyaz C., and Tamura K. ( 2018 ). MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Molecular Biology and Evolution 35 : 1547-1549.
5.https: //www.ncbi.nlm.nih.gov/pmc/articles/PMC4681159/
6. Tajima F. and Nei M. ( 1984 ). Estimation of evolutionary distance between nucleotide sequences. Molecular Biology and Evolution 1 : 269-285.