
Let's heat metal plates on the GPU
Everyone knows that Python does not shine with speed by itself. In my opinion, the language is excellent in its readability, but the main niche of its application is where you wait most of the time for some data input / output. By convention, you can write super-performing code in Rust or C, but 99% of the time it will just wait.
However, Python is also great as a high-level syntactic glue. In this case, its leisurely interpreted part invokes high-speed code written in compiled programming languages. Typically, traditional libraries such as NumPy are used for this.
But we will go a little further, try to parallelize computations in CUDA and use a strange but working hybrid of C ++, stdpar and the nvc ++ compiler from Nvidia. Well, at the same time, let's try to evaluate the performance. Let's take two problems: sorting numbers and the Jacobi method, which we will use to calculate the heating of a metal plate.
Calling C ++ from Python
Our sorting code will look like this:
# distutils: language=c++
from libcpp.algorithm cimport sort
def cppsort(int[:] x):
sort(&x[0], &x[-1] + 1)
In the first line, we explicitly indicate that Cython should use C ++, and not the default C. In the second, we import the sort function from C ++, and then the logic itself follows. Place the code in the cppsort.pyx file. Note that the extension is different from the usual py, as we will compile it or execute cythonize in Cython terminology.
Compilation can be done manually or included in setup.py, where we fully describe the preparation of our environment.
In setup.py, it looks like this:
from setuptools import setup
from Cython.Build import cythonize
setup(
ext_modules = cythonize("cppsort.pyx")
)
But we can just do this on the command line:
cythonize -i cppsort.pyx
Under the hood, something like this will happen:

- Cython will translate python code to C ++ and generate a valid cppsort.cpp.
- C ++ compiler (in this case g ++) compiles C ++ code into Python extension module
- The Python extension module is imported into the code as a regular Python module.
After compilation, we can import and immediately test the sorting:
from cppsort import cppsort
import numpy as np
x = np.array([4, 3, 2, 1], dtype="int32")
print(x)
cppsort(x)
print(x)
The array [4, 3, 2, 1] is successfully sorted to [1, 2, 3, 4] using C ++ std :: sort.
Let's go to the GPU?
C ++ standard library algorithms can be called with the parallel execution policy argument . This argument tells the compiler that you want to split the algorithm into parallel processes.
That being said, C ++ has several options for this policy:
- std :: execution :: seq: Sequential execution. Parallelism is prohibited.
- std :: execution :: unseq: Vectorized execution within the calling thread.
- std :: execution :: par: Parallel execution on one or more threads.
- std :: execution :: par_unseq: Parallel execution on one or more threads. Each stream will be vectorized whenever possible.
In this case, you yourself must monitor the race condition and deadlock. The standard g ++ compiler will try to distribute computation across CPU cores. But we can take a proprietary compiler from Nvidia - nvc ++ and feed it the "-stdpar" option. stdpar is Nvidia's C ++ Standard Parallelism with parallel code execution on the GPU.
Let's rewrite the code, taking into account the need to create a local copy of the array, since the GPU will not be able to access the array located within NumPy.
from libcpp.algorithm cimport sort, copy_n
from libcpp.vector cimport vector
from libcpp.execution cimport par
def cppsort(int[:] x):
cdef vector[int] temp
temp.resize(len(x))
copy_n(&x[0], len(x), temp.begin())
sort(par, temp.begin(), temp.end())
copy_n(temp.begin(), len(x), &x[0])

Now this needs to be compiled again, but using nvc ++. This time, let's write a normal setup.py and call it:
CC=nvc++ python setup.py build_ext --inplace
We import into the code and try to call:
x = np.array([4, 3, 2, 1], dtype="int32")
cppsort(x) # GPU
Performance
Traditionally, GPUs are good where there is a lot of the same type of lightweight computing. Heavy tasks, single GPU tasks will not work. Moreover, it is worth considering the volume of your data. If you have little data, then you will get a large overhead for the parallelization process, I / O between the CPU and GPU. As a result, such code will most likely run most efficiently on a bare CPU, sometimes even within a single core, if there is very little data. But on large arrays, the GPU will definitely be ahead.
There is a great comparison of sorting here. The NumPy speed was taken as a unit, and then the multiplicity of the speed increase in each method was calculated relative to it.
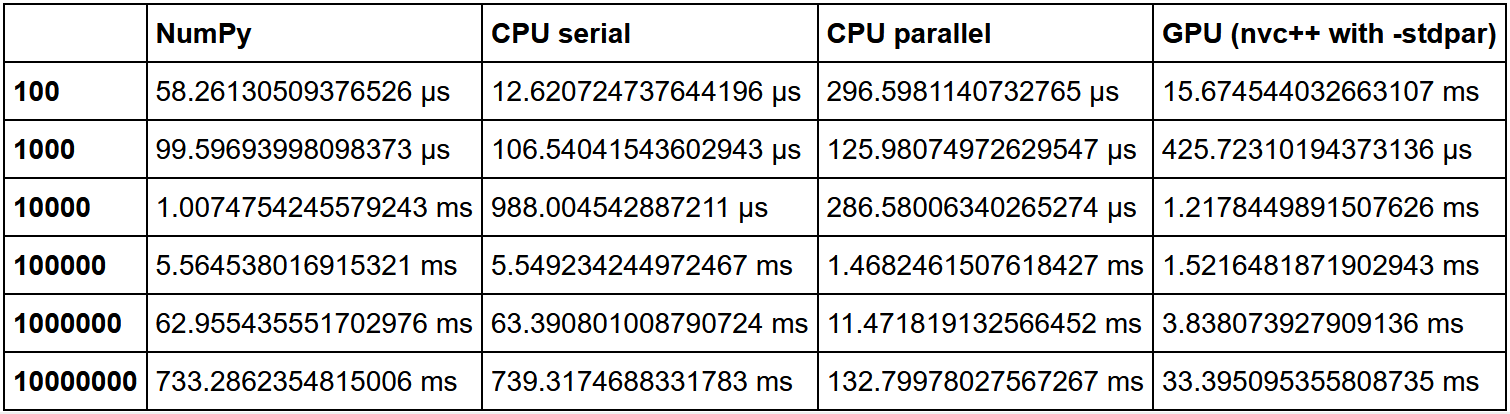
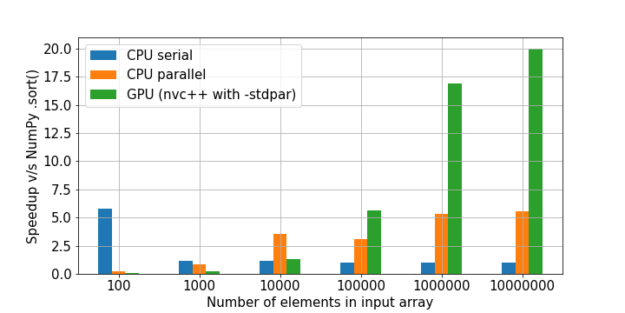
As we can see, the hypothesis that the gain in using the GPU on a large amount of data is growing is confirmed.
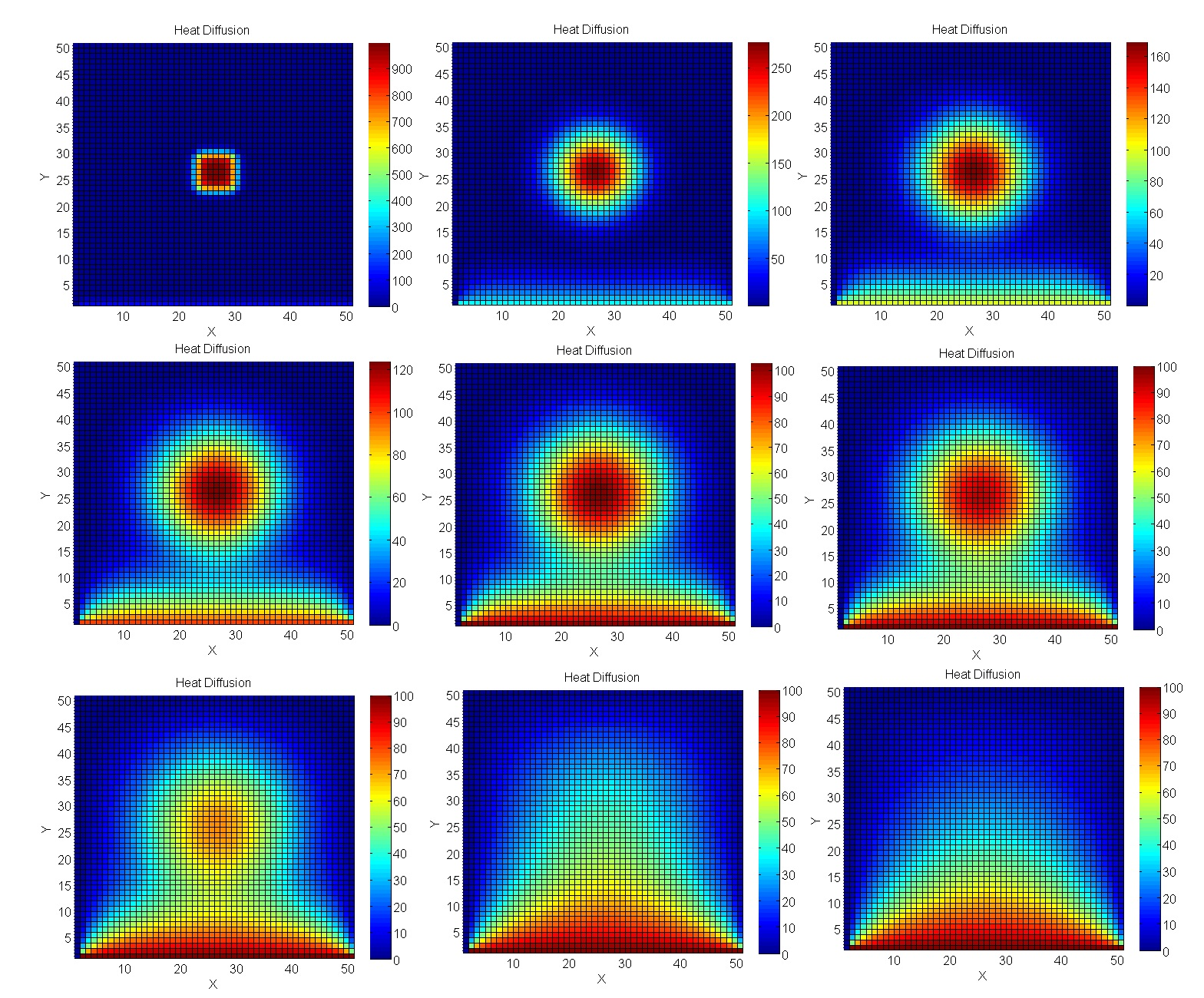
We calculate the heating of the plate
Let's take a problem closer to real engineering modeling - calculations using the Jacobi method. In particular, they are excellent for calculating temperature processes in 2D space.

Simulation code
"""
simulates heat equation on rectangle returning a heat map at a number of times
boundary and initial conditions are 0, source represents burner on a stove
This program is based on the script FEniCS tutorial demo program: Diffusion of a Gaussian hill.
u'= Laplace(u) + f in a square domain
u = u_D = 0 on the boundary
u = u_0 = 0 at t = 0
u_D = f = stove burner flame
This program succesfully runs in the fenics docker image, see the book Solving PDEs in Python.
to animate: convert -delay 4 -loop 100 heatequation10*.png heatstovelinn.gif
to crop:convert heatstovelinn.gif -coalesce -repage 0x0 -crop 810x810+95+15 +repage heatstovelin.gif
"""
from fenics import *
import time
import matplotlib.pyplot as plt
from matplotlib import cm
# Create mesh and define function space
nx = ny = 100
mesh = RectangleMesh(Point(-2, -2), Point(2, 2), nx, ny)
V = FunctionSpace(mesh, 'P', 1)
# Define boundary, source, initial
def boundary(x, on_boundary):
return on_boundary
bc = DirichletBC(V, Constant(0), boundary)
u_0 = interpolate(Constant(0), V)
f = Expression('exp(-sqrt(pow((a*pow(x[0], 2) + a*pow(x[1], 2)-a*1),2)))', degree=2, a=5) #steep guassian centred on the unit sphere
final_time = 0.035
num_pics = 72
for i in range(num_pics):
T = final_time*(i+1.0)/(num_pics+1) #solve time even space
#T = final_time*1.1**(i-num_pics+1) #solve time log space
num_steps = 30
dt = T / num_steps # time step size
# Define variational problem
u = TrialFunction(V)
v = TestFunction(V)
F = u*v*dx + dt*dot(grad(u), grad(v))*dx - (u_0 + dt*f)*v*dx
a, L = lhs(F), rhs(F)
# Time-stepping
u = Function(V)
t = 0
for n in range(num_steps):
t += dt #step
solve(a == L, u, bc) #solve
u_0.assign(u) #update
#plot solution
plot(u,cmap=cm.hot,vmin=0,vmax=0.07)
plt.axis('off')
plt.savefig('heatequation10%s.png'%(i+10),figsize=(8, 8), dpi=220,bbox_inches='tight', pad_inches=0,transparent=True)
Let's write a similar solver in Cython for subsequent compilation using CUDA:
# distutils: language=c++
# cython: cdivision=True
from libcpp.algorithm cimport swap
from libcpp.vector cimport vector
from libcpp cimport bool, float
from libc.math cimport fabs
from algorithm cimport for_each, any_of, copy
from execution cimport par, seq
cdef cppclass avg:
float *T1
float *T2
int M, N
avg(float* T1, float *T2, int M, int N):
this.T1, this.T2, this.M, this.N = T1, T2, M, N
inline void call "operator()"(int i):
if (i % this.N != 0 and i % this.N != this.N-1):
this.T2[i] = (
this.T1[i-this.N] + this.T1[i+this.N] + this.T1[i-1] + this.T1[i+1]) / 4.0
cdef cppclass converged:
float *T1
float *T2
float max_diff
converged(float* T1, float *T2, float max_diff):
this.T1, this.T2, this.max_diff = T1, T2, max_diff
inline bool call "operator()"(int i):
return fabs(this.T2[i] - this.T1[i]) > this.max_diff
def jacobi_solver(float[:, :] data, float max_diff, int max_iter=10_000):
M, N = data.shape[0], data.shape[1]
cdef vector[float] local
cdef vector[float] temp
local.resize(M*N)
temp.resize(M*N)
cdef vector[int] indices = range(N+1, (M-1)*N-1)
copy(seq, &data[0, 0], &data[-1, -1], local.begin())
copy(par, local.begin(), local.end(), temp.begin())
cdef int iterations = 0
cdef float* T1 = local.data()
cdef float* T2 = temp.data()
keep_going = True
while keep_going and iterations < max_iter:
iterations += 1
for_each(par, indices.begin(), indices.end(), avg(T1, T2, M, N))
keep_going = any_of(par, indices.begin(), indices.end(), converged(T1, T2, max_diff))
swap(T1, T2)
if (T2 == local.data()):
copy(seq, local.begin(), local.end(), &data[0, 0])
else:
copy(seq, temp.begin(), temp.end(), &data[0, 0])
return iterations
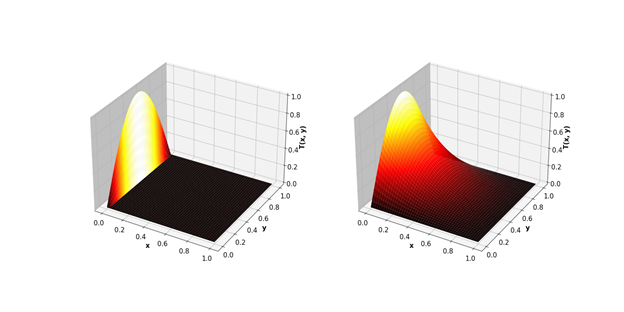

As a result, the GPU gap is even more significant.
Minuses
- Writing this kind of code is somewhat more complicated than a pure Python version and requires an understanding of how parallel computing works on a GPU.
- It is required to copy the data into a separate array for transfer to the GPU, where the video card does not have access. This can be a problem when dealing with very large arrays.
