- Allows you to write code in a familiar language, but at the same time use functions that exist only in another language.
- Allows direct collaboration with a colleague who is programming in another language.
- It makes it possible to work with two languages and eventually learn to be fluent in them.

What do we need
To work, you need these components:
- R and Python, of course.
- IDE RStudio (you can do this in other IDEs, but in RStudio it's easier).
- Your favorite Python environment manager (I'm using conda here).
- Packages
rmarkdown
andreticulate
installed in R.
When writing R Markdown documents, we will be working in RStudio, but at the same time navigate between code snippets written in R and in Python. I'll show you a couple of simple examples.
Setting up the Python environment
If you are familiar with Python programming, then you know that any work done in Python must refer to a specific environment that contains all the packages necessary for the work. There are many ways to manage packages in Python, the two most popular are virtualenv and conda. Here I am assuming that we are using conda and that it is installed as the Python environment manager.
You can use the reticulate package in R to set up conda environments via the R command line if you like (using features like
conda_create()
), but as a regular Python programmer, I prefer to set up my environments manually.
Suppose we create a conda environment named
r_and_python
and install into it
pandas
and
statsmodels
... So the commands in the terminal:
conda create -name r_and_python conda activate r_and_python conda install pandas conda install statsmodels
After installing
pandas
,
statsmodels
(and any other packages you may need), the environment setup is complete. Now run conda info in terminal and select the path to your environment. You will need it in the next step.
Setting up your R project to work with R and Python
We will start an R project in RStudio, but we want to be able to run Python in the same project. To ensure that the Python code runs in the environment we want, we need to set the system environment variable
RETICULATE_PYTHON
for the Python executable in that environment. This will be the path you chose in the previous section, followed by
/bin/python3
.
The best way to ensure that this variable is permanently set in your project is to create a text file named in the project
.Rprofile
and add this line to it.
Sys.setenv(RETICULATE_PYTHON=”path_to_environment/bin/python3")
Replace pathtoenvironment with the path you chose in the previous section. Save the file
.Rprofile
and restart the R session. Each time you restart a session or project, it starts up
.Rprofile
, setting up your Python environment. If you want to test this, you can run the line Sys.getenv ("RETICULATE_PYTHON").
Writing Code - First Example
Now you can set up an R Markdown document in your project
.Rmd
and write code in two different languages. First you need to load the reticulate library in your first piece of code.
```{r} library(reticulate) ```
Now, when you want to write Python code, you can wrap it with normal back quotes, but mark it as a Python code snippet with
{python}
, and when you want to write in R, use
{r}
.
For our first example, suppose you run a Python model on a dataset of student test scores.
```{python} import pandas as pd import statsmodels.api as sm import statsmodels.formula.api as smf # obtain ugtests data url = “http://peopleanalytics-regression-book.org/data/ugtests.csv" ugtests = pd.read_csv(url) # define model model = smf.ols(formula = “Final ~ Yr3 + Yr2 + Yr1”, data = ugtests) # fit model fitted_model = model.fit() # see results summary model_summary = fitted_model.summary() print(model_summary) ```
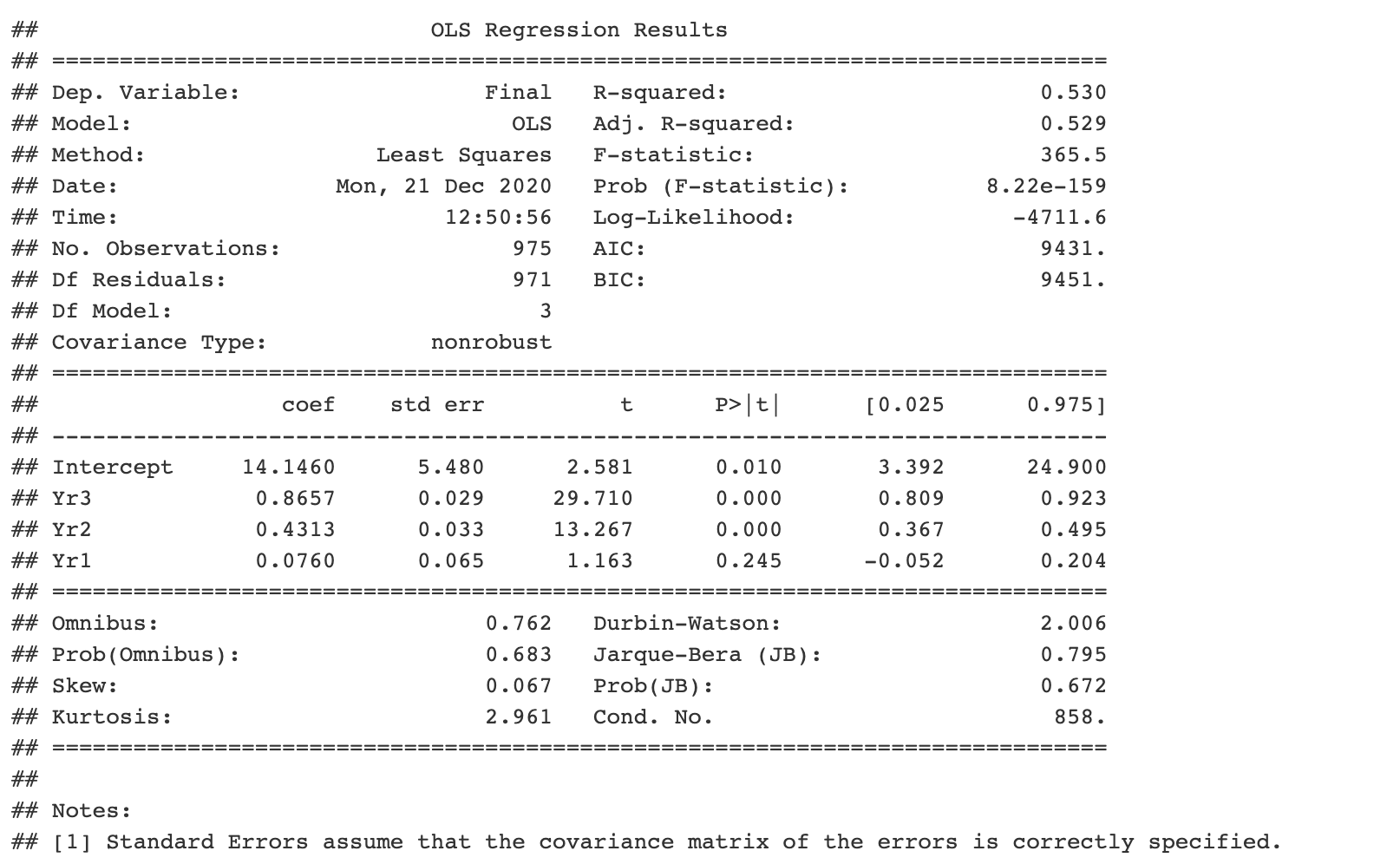
That's great, but let's say you had to quit your job because of something more urgent and hand it over to your colleague, the R programmer. You were hoping that you could diagnose the model.
Do not be afraid. You can access all the python objects that you have created in the general list called py. So if an R block is created inside your R Markdown document, colleagues will have access to your model parameters:
```{r} py$fitted_model$params ```

or the first few leftovers:
```{r} py$fitted_model$resid[1:5] ```

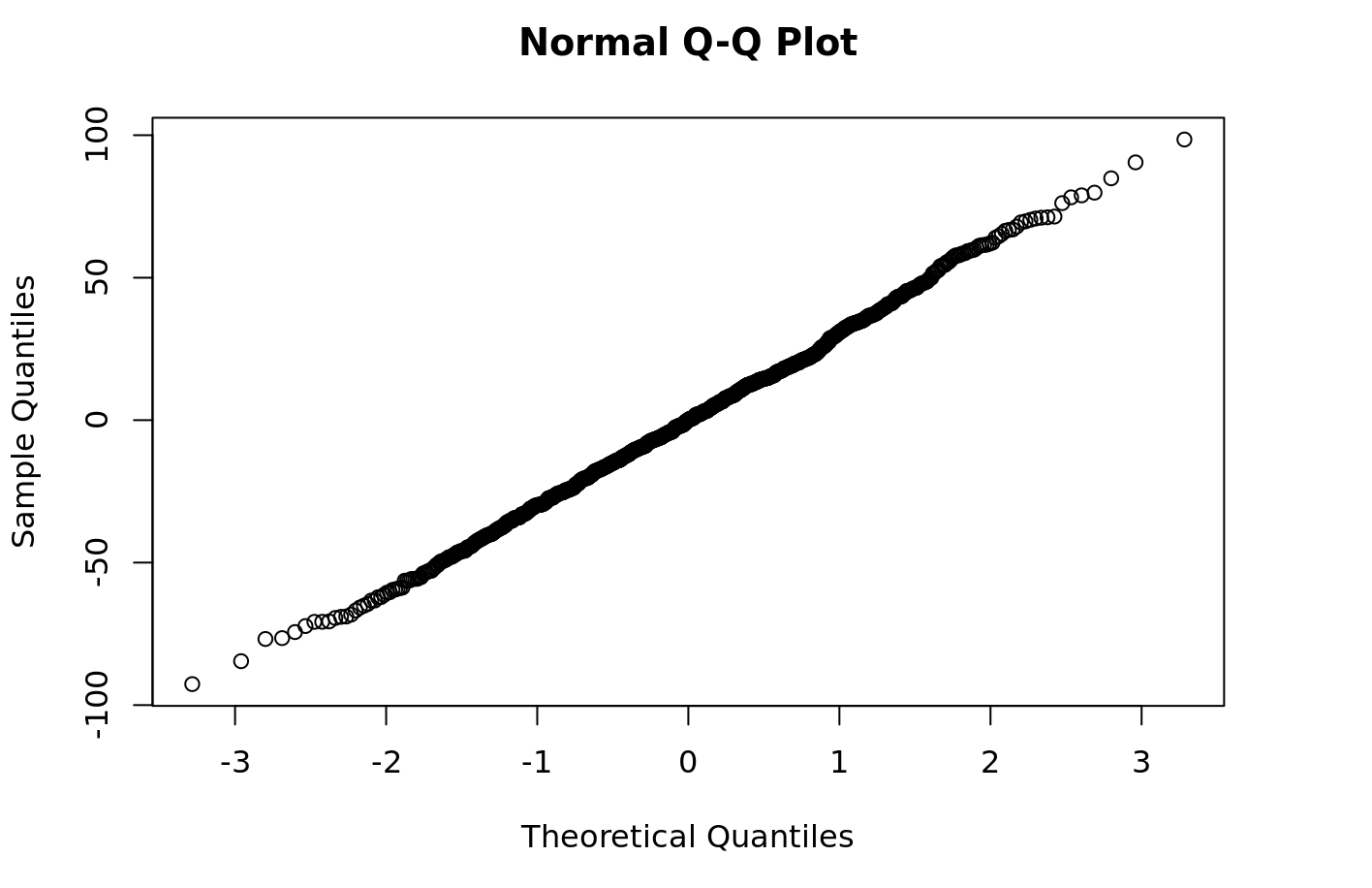
Now you can easily perform some diagnostics on the model, such as plotting the residuals of your quantile-quantile model:
```{r} qqnorm(py$fitted_model$resid) ```
Writing code - second example
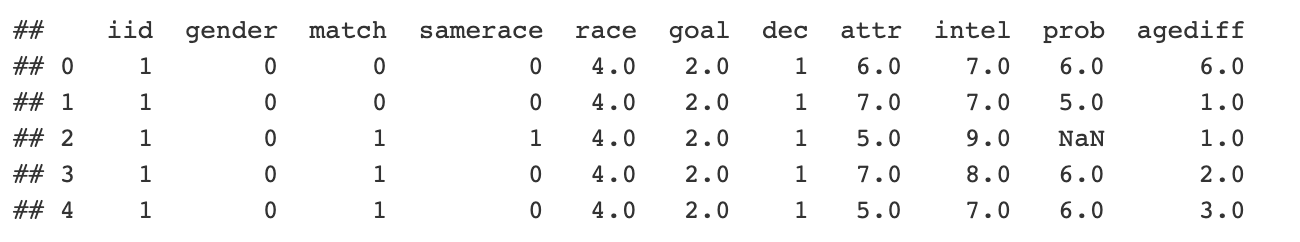
You parsed some Python dating data and created a pandas dataframe with all the data in it. For simplicity, let's load the data and look at it:
```{python} import pandas as pd url = “http://peopleanalytics-regression-book.org/data/speed_dating.csv" speed_dating = pd.read_csv(url) print(speed_dating.head()) ```
You have now run a simple logistic regression model in Python to try and associate the dec solution with some other variables. However, you understand that this data is actually hierarchical and that the same individual iid can have multiple acquaintances.
So you know you need to run a mixed-effects logistic regression model, but you can't find any Python program that does it!
And again, don't be afraid, send the project to a colleague and he will write the solution in R.
```{r} library(lme4) speed_dating <- py$speed_dating iid_intercept_model <- lme4:::glmer(dec ~ agediff + samerace + attr + intel + prob + (1 | iid), data = speed_dating, family = “binomial”) coefficients <- coef(iid_intercept_model)$iid ```
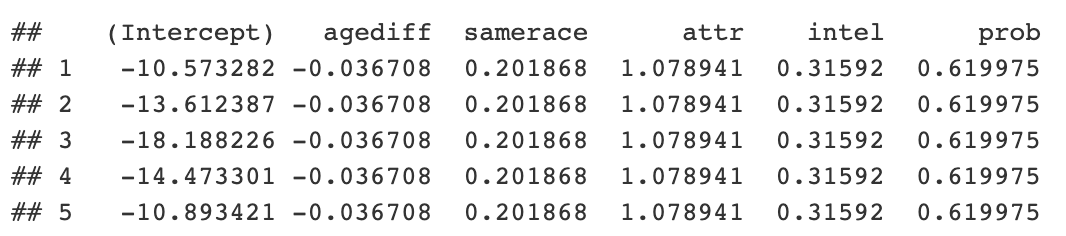
Now you can get the code and look at the odds. It is also possible to access Python R objects inside a generic r object.
```{python} coefs = r.coefficients print(coefs.head()) ```
These two examples show how you can seamlessly navigate between R and Python in the same R Markdown document. So the next time you think about working on a cross-language project, think about running all the steps in R Markdown. This can save you a lot of the hassle of switching between two languages and help keep all of your work in one place as a continuous narrative.
You can see the finished R Markdown document built around language integration - with snippets of R and Python and objects moving between them - posted here . The Github repository with the source code is here .
The sample data in the document is from my The People Analytics Regression Modeling Reference .

Other professions and courses
PROFESSION
COURSES
- Java developer profession
- Frontend developer profession
- Profession Web developer
- Profession Ethical hacker
- C ++ developer profession
- Profession Unity Game Developer
- The profession of iOS developer from scratch
- Profession Android developer from scratch
COURSES
- Machine Learning Course
- Advanced Course "Machine Learning Pro + Deep Learning"
- Python for Web Development Course
- JavaScript course
- « Machine Learning Data Science»
- DevOps