
Left - Tanay Tandon, at the age of 17 in 2014, founded the startup Athelas to diagnose malaria using a smartphone. Right: Athelas co-founder Deepika Bodopati.
Several years ago, Athelas (YC S16) developed its prototype overnight at the YC Hacks 2014 hackathon . This month, we began shipping the device to hospitals and patients across the country. During this time, we have learned a lot and would like to share some thoughts.

Athelas is an inexpensive imager that provides instant blood analysis using computer vision instead of traditional laboratory tests. The path from a prototype assembled on the knee to a device ready for delivery (especially in medicine) at every stage was a change in dimension, and now it is interesting to remember the very first day.
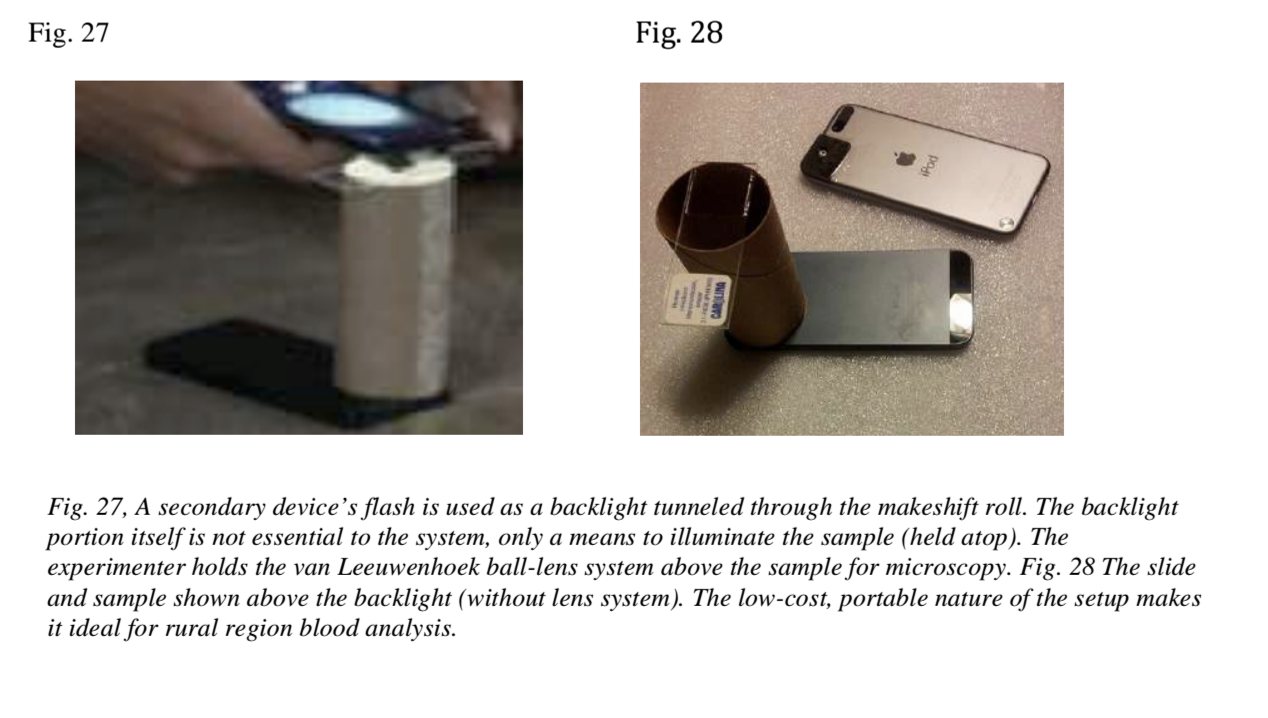
The version that was assembled at the hackathon used a piece of foam rubber and a spherical magnifier attached to the smartphone's camera. The blood sample was held underneath (in a roll of toilet paper), the camera took several pictures, and then the malaria cells were counted using computer vision. The design of the device is very similar to the van Leeuwenhoek microscope(considered one of the very first microscopes), which was used for the first time in human history to observe microorganisms. There have been several attempts to put everything together, and I spent a couple of hours of hackathon getting the device to work reliably with my phone.


Fragments of notes that I took a few months after the hackathon.
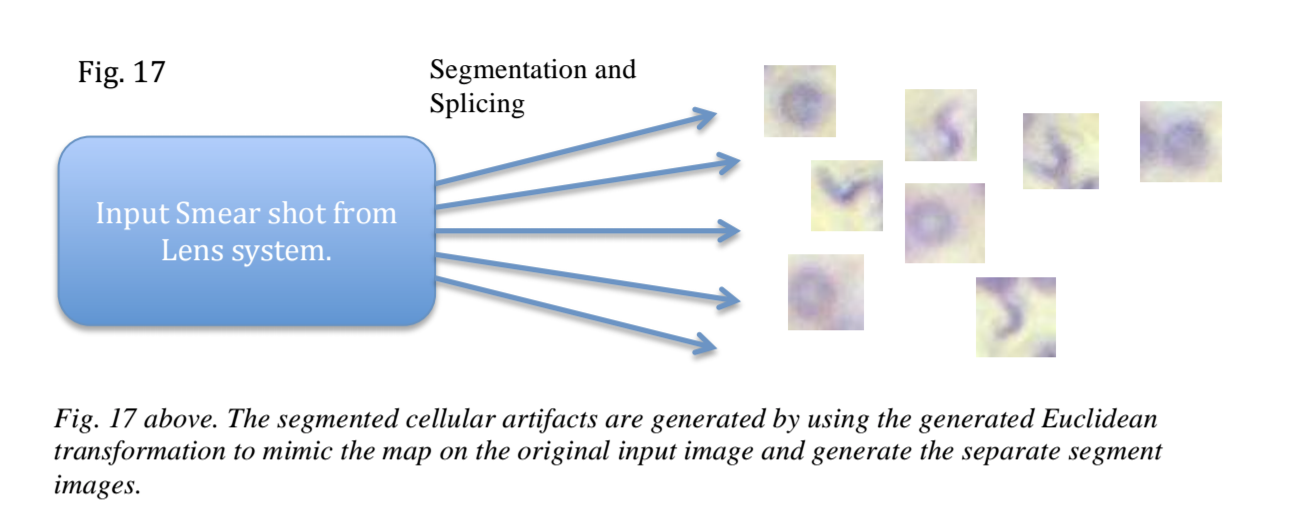
The real hack was segmentation and pattern matching combined with an implementation of the Fast Random Forest model trained to classify the extracted red blood cell (RBC) traits. Cell boundaries are recognized and then fed to a classifier to determine if there are parasitic cells (such as malaria or trypanosomes).
The prototype was made for a cool presentation, when the glasses with the blood sample show the malaria parasite, but the blood of a normal person does not. During the demonstration, someone physically had to hold the camera in one place, the glasses with the samples were replaced by a clever trick, and the light had to be caught. By the end of the day, the prototype was a fun toy to experiment with, you may have seen the Facebook video.
But they were sure that it was something more. The key idea was - if we make a widely applicable and easy-to-use device, why not do a simple blood test in every doctor's office, examination room, or even at home? Upon returning home, this idea completely absorbed us, and we decided to develop it - but as a product, and not just a prototype. This meant the creation of an automated mechanism for analyzing peripheral blood smears, a more reliable computer vision approach for various cell types, automated mechanisms for imaging the entire sample without storing slides, and, more importantly, conducting clinical validation studies.

Deepika (my co-founder) worked to improve the method for quickly staining cells and came up with a way to apply the dye to plastic strips that you can use right out of the box. She worked mainly in the laboratory, synthesizing dozens of dye variants and conducting empirical studies of the quality of cell display. Another part of the problem was the need to lightly compress the strip to create a "monolayer" or single layer of cells that allows statistically representative images to be obtained.


Excerpts from "The marching velocity of the capillary meniscus in a microchannel", the example we are referring to when simulating channel flow to create a "monolayer". This capillary development was ultimately put on hold.
In the meantime, I focused on creating high-resolution optics for a still inexpensive but self-contained device. Thus, we could focus on monitoring the prevailing cell types such as white blood cells and platelets (other than malaria). At the heart of it all was the triggering system, which combined Gaussian image focusing algorithms to ensure robust cell recognition. Here's a prototype halfway through:

In addition, we began building a training sample of CDC (Centers for Disease Control and Prevention) and blood smears collected by researchers at Stanford and UCLA - often manually tagged by me or a pathologist. From that point on, we were able to use traditional computer vision and deep learning approaches to recognize and classify cell types based on previous human verified examples.

Image of the extracted cell bodies after Hough transformation, first pass, preliminary segmentation and classification
The first steps were hard. The Stanford college workload + rising hardware costs have reduced the iteration speed required for normal product development. Finals often meant days passed without any noticeable progress. But we put together a tactile, usable v1 that can capture and process an image of a stained blood sample. See demo:
This summer in this orange building in Mountain View, things got back to normal when we joined Y Combinator's summer batch. All of our efforts (and now we have a full-time project) have been focused on clinical validation at the FEMAP Family Clinic to launch the first series of applications within the healthcare system. The goal was to test the system in only one aspect: leukocyte counting. By capturing images of blood samples on our strip and then running algorithms, we showed how our highly correlated cell counts for 350 patients matched the gold standard of Beckman Coulter's cell counters, combined with a series of laboratory accuracy checks .

An interesting aspect was that the drop-to-drop accuracy we showed (which has attracted increased interest in recent years ) has been clinically confirmed in comparison with other systems using blood drops. Coulter counters (traditional cell counting systems) work by passing particles through a precious aperture a few microns in diameter and capturing the characteristic impedance to record the particle size and thus its classification. Basically, the higher the impedance, the larger the particle size .
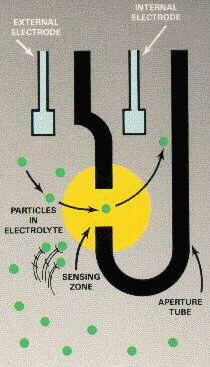
Coulter counter principle schematic, source: cyto.purdue.edu.
However, Athelas' approach to computer vision focuses entirely on imaging and nucleation models. Therefore, particulate matter or lymph, which can often confuse the Coulter system (especially in dilute amounts), is simply classified by computer vision as a non-leukocyte cell body - not a leukocyte, but some other unclassified artifact in a blood sample.

The test has shown a high degree of agreement between experts (100% agreement between experts on Grade 5) between the two systems, we have filed documents with the FDA (Food and Drug Administration) to register the system under Class 2 510 (k), we are now selling the Class 1 version of the system for rapid monitoring of leukocytes. Learn more at athelas.com.

In the coming months, we will integrate new types of blood tests into the system (monitoring for concussion, tracking inflammation, urinary tract infections, platelets, increasing the number of cells), our key task will be to interact with the professional medical community in order to accept and implement our system.

At the same time, we will focus on shipping our $ 250 devices to as many outlets as possible, healthcare facilities and households.
We are constantly looking for outstanding people to meet and hackers to join our team, so write me if you want to chat: tanay [at] getathelas.com
Translation: Ilya Lankevich
If you want to help with translations of useful materials from the YC library, write in a personal, @jethacker cart or mail alexey.stacenko@gmail.com
Follow the YC Startup Library news in Russian in the telegram channel or on Facebook .