We will tell you why this tool appeared and what it can do.

Lack of algorithms
One of the key challenges in machine learning is data dimensionality reduction. Data Scientists reduce the number of variables by isolating among them the values that have the greatest impact on the result. After this operation, the machine learning model requires less memory, works faster and better. The example below shows that eliminating duplicate features increases the classification accuracy from 0.903 to 0.943.
>>> from sklearn.linear_model import SGDClassifier
>>> from ITMO_FS.embedded import MOS
>>> X, y = make_classification(n_samples=300, n_features=10, random_state=0, n_informative=2)
>>> sel = MOS()
>>> trX = sel.fit_transform(X, y, smote=False)
>>> cl1 = SGDClassifier()
>>> cl1.fit(X, y)
>>> cl1.score(X, y)
0.9033333333333333
>>> cl2 = SGDClassifier()
>>> cl2.fit(trX, y)
>>> cl2.score(trX, y)
0.9433333333333334There are two approaches to dimensionality reduction — feature design and feature selection. In fields like bioinformatics and medicine, the latter is often used, since it allows you to highlight significant features while preserving semantics, that is, it does not change the original meaning of features. However, the most common Python machine learning libraries - scikit-learn, pytorch, keras, tensorflow - lack a complete set of feature selection methods.
To solve this problem, ITMO University students and postgraduates have developed an open library - ITMO_FS. A team is working on it under the leadership of Ivan Smetannikov, Associate Professor of the Faculty of Information Technologies and Programming, Deputy Head of the Machine Learning Laboratory. Lead developer - Nikita Pilnenskiy, graduated from the Master's degree in Machine Learning and Data Analysis . Now he goes to graduate school.
« , . , , , (-) .
, , , . , , , ».
—
ITMO_FS is implemented in Python and is compatible with scikit-learn, which is considered the de facto main data analysis tool. Its feature selectors take the same parameters:
data: array-like (2-D list, pandas.Dataframe, numpy.array);
targets: array-like (1-D list, pandas.Series, numpy.array).The library supports all classic approaches to feature selection - filters, wrappers and inline methods. Among them are such algorithms as filters based on Spearman and Pearson correlations, Fit Criterion, QPFS, hill climbing filter and others .

The library also supports training ensembles by combining feature selection algorithms based on the measures of significance used in them. This approach allows you to obtain higher predictive results with a low time investment.
What are the analogues
There are not many feature selection algorithms libraries, especially in Python. One of the largest is considered the development of engineers from the Arizona State University (ASU). It supports a large number of algorithms, but has hardly been updated recently.
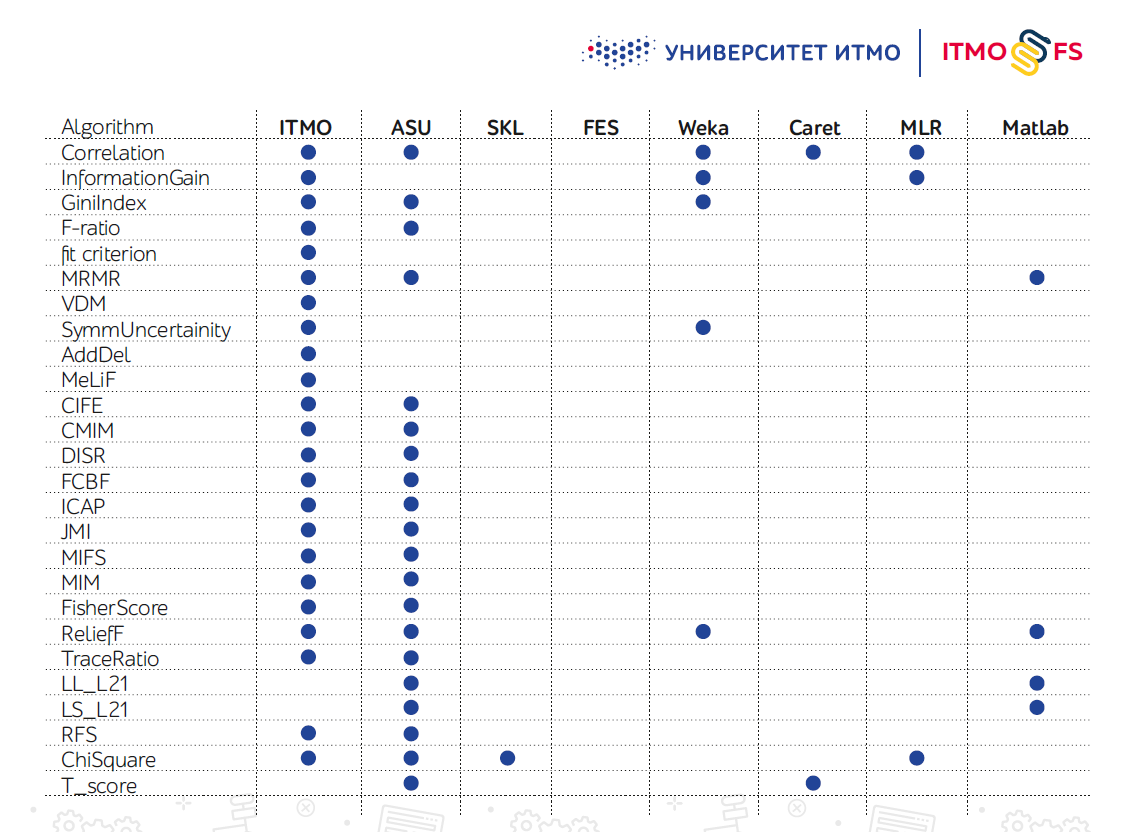
Scikit-learn itself also has several feature selection mechanisms, but in practice they are not enough.
"In general, over the past five to seven years, the focus has shifted towards ensemble algorithms for feature selection, but they are not particularly represented in such libraries, which we also want to fix."
- Ivan Smetannikov
Project prospects
The authors of ITMO_FS plan to integrate their product with scikit-learn by adding it to the list of officially compatible libraries. At the moment, the library already contains the largest number of feature selection algorithms among all libraries, but their addition continues. Further on the roadmap is the addition of new algorithms, including our own developments.
In more distant plans, there are tasks to introduce the library into the meta-learning system, add algorithms for direct work with matrix data (filling in gaps, generating meta-attribute space data, etc.), as well as a graphical interface. In parallel with this, hackathons will be held using the library in order to interest more developers in the product and get feedback.
It is expected that ITMO_FS will find application in the fields of medicine and bioinformatics - in such problems as the diagnosis of various cancers, the construction of predictive models of phenotypic characteristics (for example, a person's age) and the synthesis of drugs.
Where can I download
If you are interested in the ITMO_FS project, you can download the library and try it out in practice - here is the repository on GitHub . An initial version of the documentation is available at readthedocs . There you can also see the installation instructions (supported by pip). We welcome any feedback.
Additional materials from our blog on Habré:
- Podcast: what awaits aspiring scientists in ML
- Podcast: Quantum Hacking and Key Sharing