But the work of Data Scientist is tied to data, and one of the most important and time-consuming moments is processing data before submitting it to a neural network or analyzing it in a certain way.
In this article, our team will describe how you can quickly and easily process data with step-by-step instructions and code. We tried to make the code flexible enough to be applied across different datasets.
Many professionals may not find anything extraordinary in this article, but beginners will be able to learn something new, and anyone who has long dreamed of making a separate notebook for fast and structured data processing can copy the code and format it for themselves, or download a ready-made one. notebook from Github.
We got dataset. What to do next?
So, the standard: you need to understand what we are dealing with, the big picture. We'll use pandas for this to simply define different data types.
import pandas as pd # pandas
import numpy as np # numpy
df = pd.read_csv("AB_NYC_2019.csv") # df
df.head(3) # 3 , , 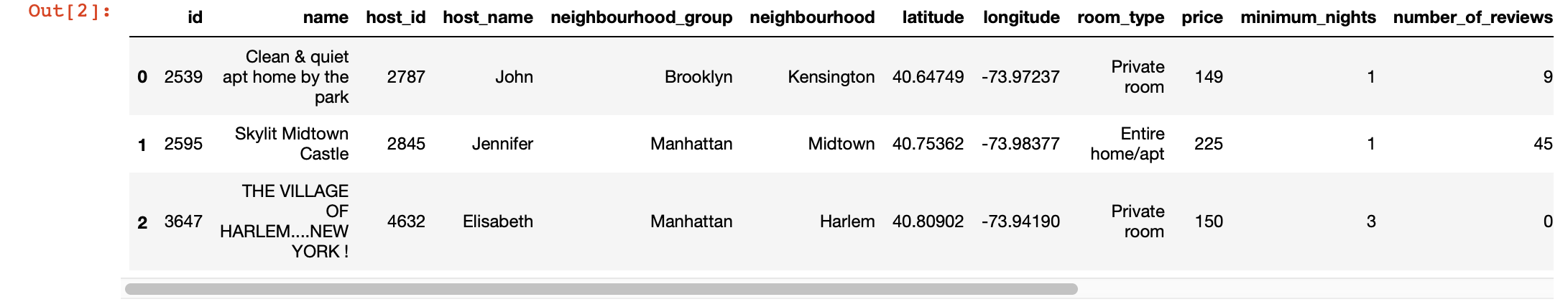
df.info() # 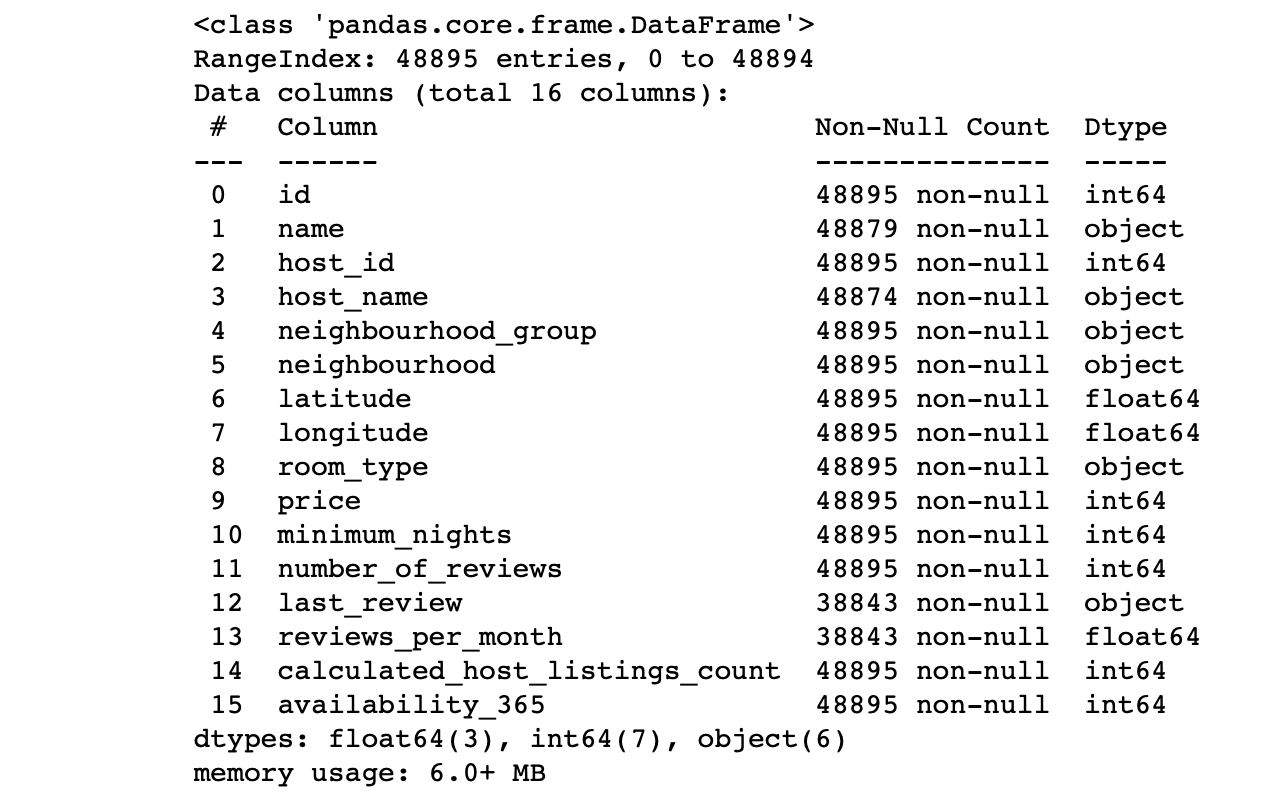
We look at the values of the columns:
- Does the number of lines in each column correspond to the total number of lines?
- What is the essence of the data in each column?
- What column do we want target to make predictions for?
The answers to these questions will allow you to analyze the dataset and roughly draw a plan for the next steps.
We can also use pandas describe () for a deeper look at the values in each column. However, the disadvantage of this function is that it does not provide information about columns with string values. We will deal with them later.
df.describe()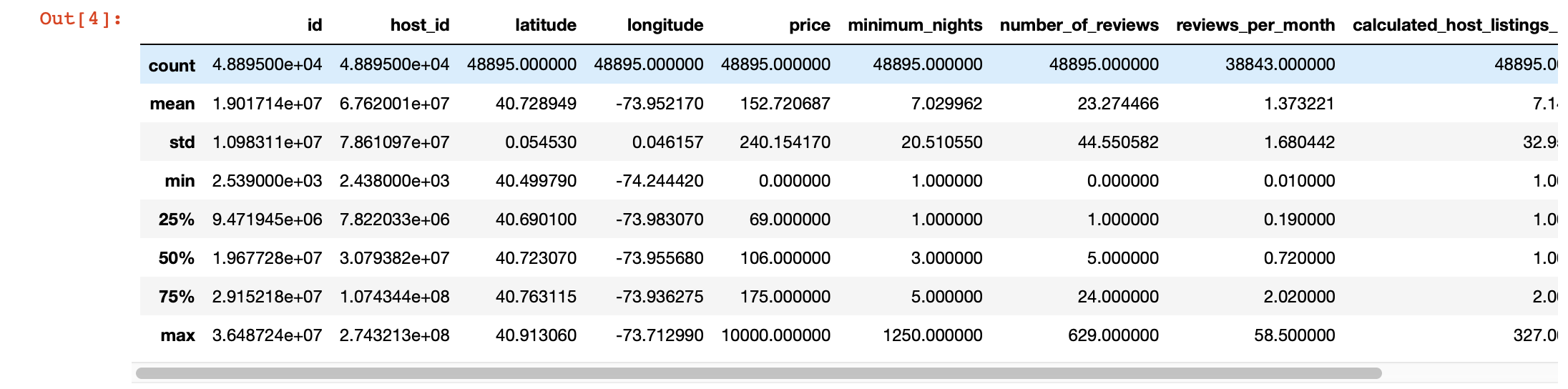
Magic visualization
Let's look at where we have no values at all:
import seaborn as sns
sns.heatmap(df.isnull(),yticklabels=False,cbar=False,cmap='viridis')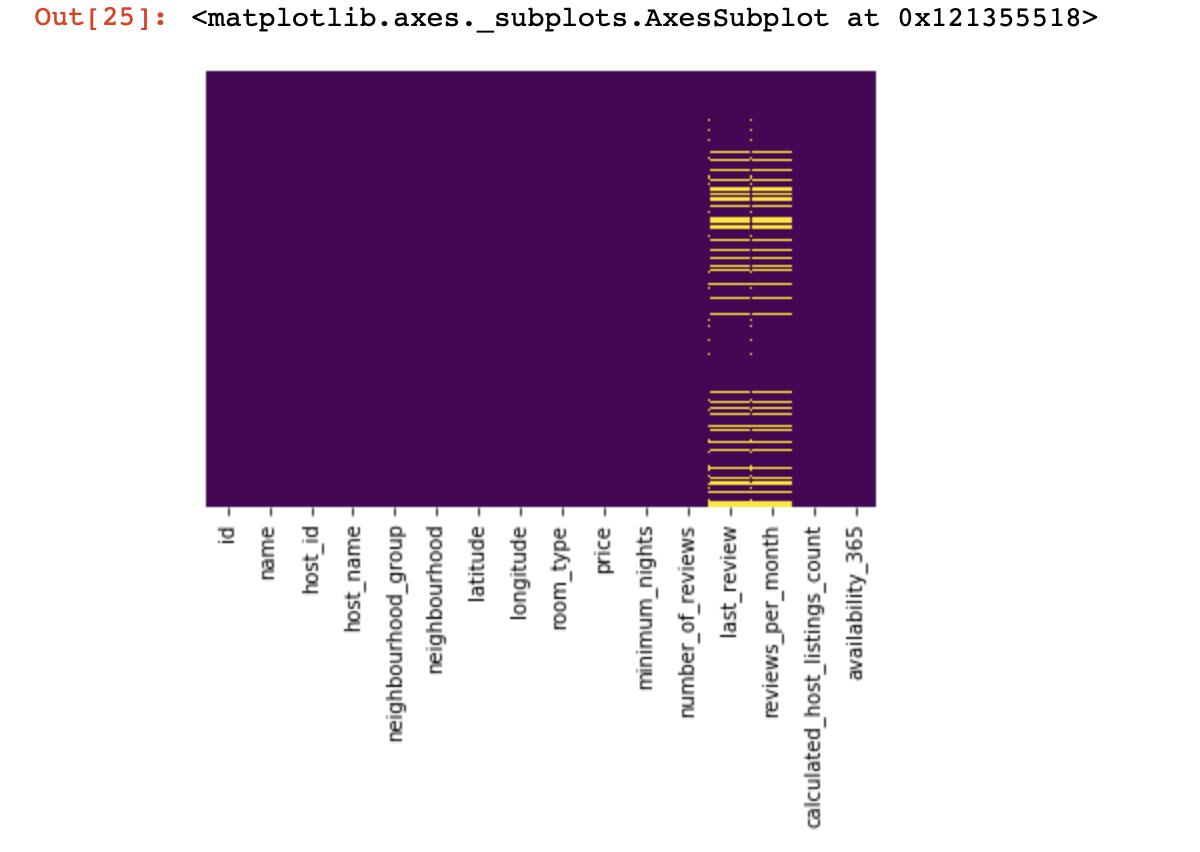
It was a small look from above, now we will proceed to more interesting things.Let's
try to find and, if possible, delete columns that have only one value in all lines (they will not affect the result in any way):
df = df[[c for c
in list(df)
if len(df[c].unique()) > 1]] # , , Now we protect ourselves and the success of our project from duplicate lines (lines that contain the same information in the same order as one of the existing lines):
df.drop_duplicates(inplace=True) # , .
# .We divide the dataset into two: one with qualitative values, and the other with quantitative values
Here we need to make a small clarification: if the lines with missing data in qualitative and quantitative data do not strongly correlate with each other, then it will be necessary to decide what we sacrifice - all lines with missing data, only part of them, or certain columns. If the lines are related, then we have every right to divide the dataset into two. Otherwise, you will first need to deal with the lines that do not correlate the missing data in qualitative and quantitative terms, and only then divide the dataset into two.
df_numerical = df.select_dtypes(include = [np.number])
df_categorical = df.select_dtypes(exclude = [np.number])We do this to make it easier for us to process these two different types of data - later we will understand how much it simplifies our life.
We work with quantitative data
The first thing we need to do is determine if there are any "spy columns" in the quantitative data. We call these columns this because they pretend to be quantitative data, and they themselves work as qualitative data.
How do we define them? Of course, it all depends on the nature of the data that you are analyzing, but in general, such columns may have few unique data (in the region of 3-10 unique values).
print(df_numerical.nunique())After we define the spy columns, we will move them from quantitative to qualitative data:
spy_columns = df_numerical[['1', '2', '3']]# - dataframe
df_numerical.drop(labels=['1', '2', '3'], axis=1, inplace = True)#
df_categorical.insert(1, '1', spy_columns['1']) # -
df_categorical.insert(1, '2', spy_columns['2']) # -
df_categorical.insert(1, '3', spy_columns['3']) # - Finally, we completely separated quantitative data from qualitative data and now you can work with them properly. The first is to understand where we have empty values (NaN, and in some cases 0 will be taken as empty values).
for i in df_numerical.columns:
print(i, df[i][df[i]==0].count())At this stage, it is important to understand in which columns the zeros can mean missing values: is this related to how the data was collected? Or could it be related to data values? These questions need to be answered on a case-by-case basis.
So, if we nevertheless decided that we may not have data where there are zeros, we should replace the zeros with NaN, so that it would be easier to work with this lost data later:
df_numerical[[" 1", " 2"]] = df_numerical[[" 1", " 2"]].replace(0, nan)Now let's see where we have missing data:
sns.heatmap(df_numerical.isnull(),yticklabels=False,cbar=False,cmap='viridis') # df_numerical.info()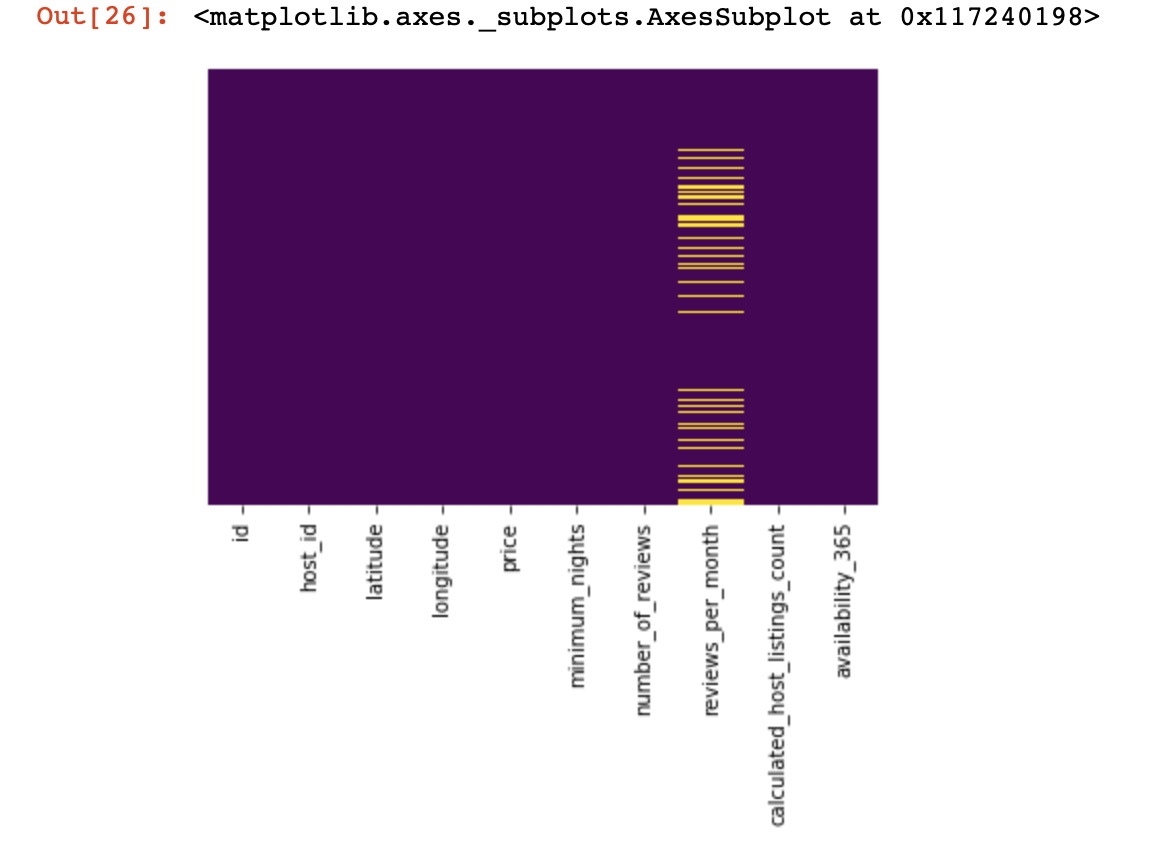
Here, those values inside the columns that are missing should be marked in yellow. And the fun begins now - how to behave with these values? Delete lines with these values or columns? Or fill these empty values with some other?
Here is a rough diagram that can help you decide what you can basically do with empty values:
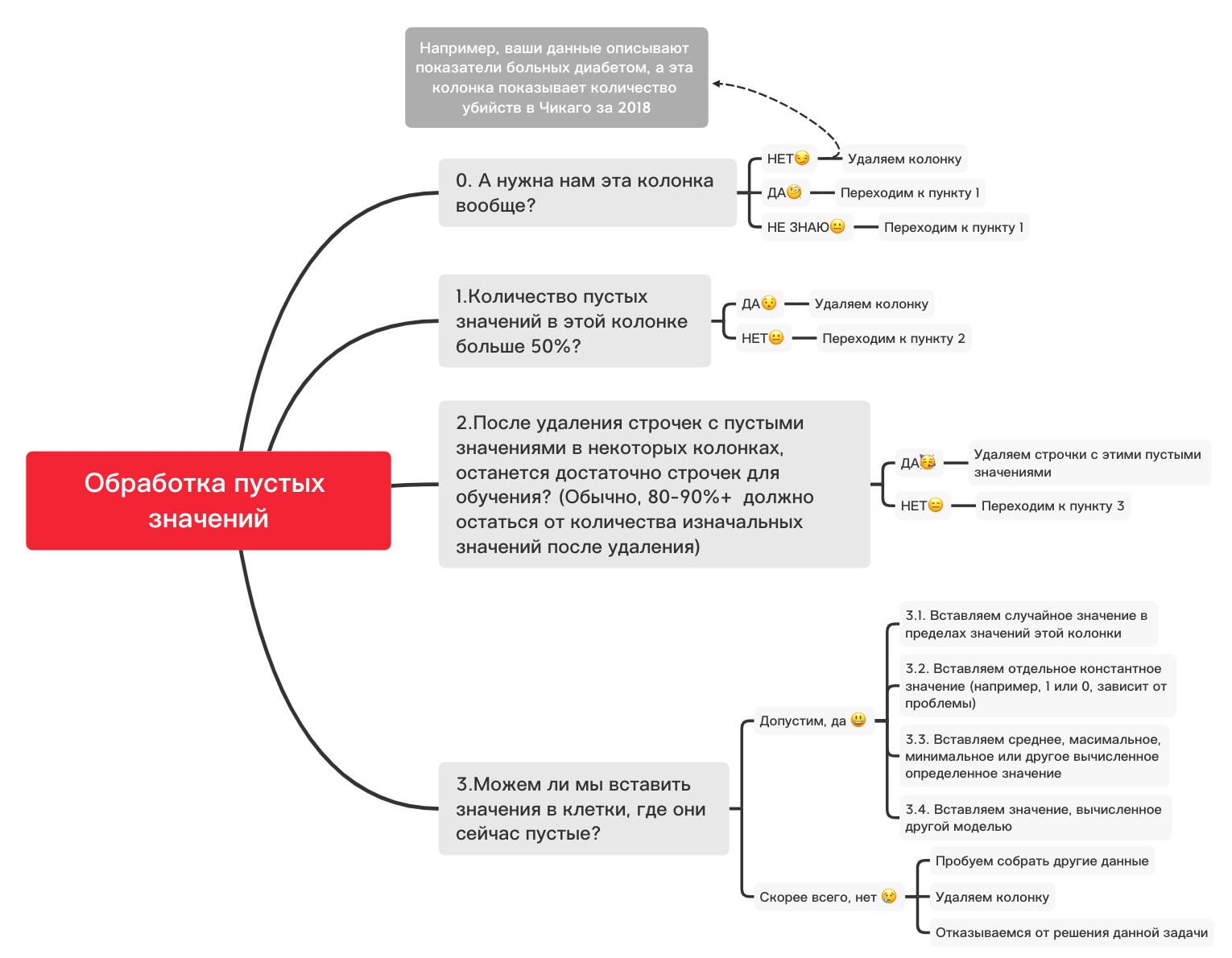
0. Remove unnecessary columns
df_numerical.drop(labels=["1","2"], axis=1, inplace=True)1. Are there more than 50% blank values in this column?
print(df_numerical.isnull().sum() / df_numerical.shape[0] * 100)df_numerical.drop(labels=["1","2"], axis=1, inplace=True)#, - 50 2. Delete lines with empty values
df_numerical.dropna(inplace=True)# , 3.1. Insert a random value
import random # random
df_numerical[""].fillna(lambda x: random.choice(df[df[column] != np.nan][""]), inplace=True) # 3.2. Insert constant value
from sklearn.impute import SimpleImputer # SimpleImputer,
imputer = SimpleImputer(strategy='constant', fill_value="< >") # SimpleImputer
df_numerical[["_1",'_2','_3']] = imputer.fit_transform(df_numerical[['1', '2', '3']]) #
df_numerical.drop(labels = ["1","2","3"], axis = 1, inplace = True) # 3.3. Insert the average or the most frequent value
from sklearn.impute import SimpleImputer # SimpleImputer,
imputer = SimpleImputer(strategy='mean', missing_values = np.nan) # mean most_frequent
df_numerical[["_1",'_2','_3']] = imputer.fit_transform(df_numerical[['1', '2', '3']]) #
df_numerical.drop(labels = ["1","2","3"], axis = 1, inplace = True) # 3.4. Inserting a Value Calculated by Another Model
Sometimes values can be calculated using regression models using models from the sklearn library or other similar libraries. Our team will devote a separate article on how this can be done in the near future.
So, while the narrative about quantitative data will be interrupted, because there are many other nuances about how to better do data preparation and preprocessing for different tasks, and basic things for quantitative data have been taken into account in this article, and now is the time to return to qualitative data. which we have separated a few steps back from quantitative. You can change this notebook as you like, adjusting it for different tasks, so that data preprocessing goes very quickly!
Qualitative data
Basically, for quality data, the One-hot-encoding method is used in order to format it from a string (or object) to a number. Before moving on to this point, let's use the diagram and the code above in order to deal with empty values.
df_categorical.nunique()sns.heatmap(df_categorical.isnull(),yticklabels=False,cbar=False,cmap='viridis')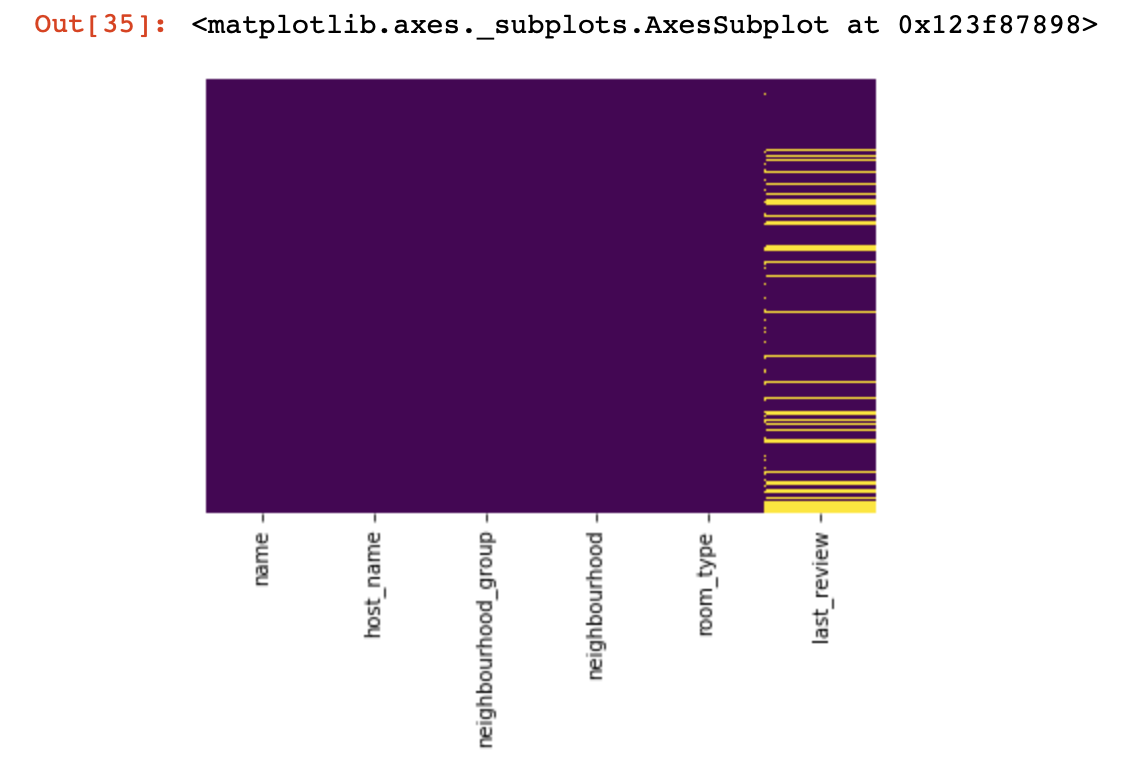
0. Removing unnecessary columns
df_categorical.drop(labels=["1","2"], axis=1, inplace=True)1. Are there more than 50% blank values in this column?
print(df_categorical.isnull().sum() / df_numerical.shape[0] * 100)df_categorical.drop(labels=["1","2"], axis=1, inplace=True) #, -
# 50% 2. Delete lines with empty values
df_categorical.dropna(inplace=True)# ,
# 3.1. Insert a random value
import random
df_categorical[""].fillna(lambda x: random.choice(df[df[column] != np.nan][""]), inplace=True)3.2. Insert constant value
from sklearn.impute import SimpleImputer
imputer = SimpleImputer(strategy='constant', fill_value="< >")
df_categorical[["_1",'_2','_3']] = imputer.fit_transform(df_categorical[['1', '2', '3']])
df_categorical.drop(labels = ["1","2","3"], axis = 1, inplace = True)So, finally, we have dealt with empty values in quality data. Now is the time to one-hot-encoding the values that are in your database. This method is very often used so that your algorithm can train with good data.
def encode_and_bind(original_dataframe, feature_to_encode):
dummies = pd.get_dummies(original_dataframe[[feature_to_encode]])
res = pd.concat([original_dataframe, dummies], axis=1)
res = res.drop([feature_to_encode], axis=1)
return(res)features_to_encode = ["1","2","3"]
for feature in features_to_encode:
df_categorical = encode_and_bind(df_categorical, feature))So, finally we have finished processing qualitative and quantitative data separately - it's time to combine them back
new_df = pd.concat([df_numerical,df_categorical], axis=1)After we have connected the datasets together into one, in the end we can use the data transformation using MinMaxScaler from the sklearn library. This will make our values between 0 and 1, which will help when training the model in the future.
from sklearn.preprocessing import MinMaxScaler
min_max_scaler = MinMaxScaler()
new_df = min_max_scaler.fit_transform(new_df)This data is now ready for anything - for neural networks, standard ML algorithms, and so on!
In this article, we did not take into account the work with data related to time series, since for such data slightly different processing techniques should be used, depending on your task. In the future, our team will devote a separate article to this topic, and we hope that it will be able to bring something interesting, new and useful into your life, like this one.